On September 27, 2019, the annual meeting of the European Society of Oncology (ESMO) kicked off in Barcelona, Spain. ESMO is Europe’s most prestigious and influential oncology conference, and many clinical institutions and leading companies have published the latest translational research and clinical practice data in the field of cancer at this annual meeting.
On September 28th, GRAIL, the US cancer screening company, released the latest research data of the company’s cfDNA-based targeted methylation liquid biopsy analysis method in the field of cancer early screening at the ESMO annual meeting. The results of this multicenter study show that by analyzing the cancer-associated methylation patterns of cfDNA, the company’s method is capable of detecting and localizing more than 20 types of cancer with high accuracy. Dr. Geoffrey R. Oxnard from the Dana-Farber Cancer Institute in the United States gave an oral report on the latest data.
GRAIL hopes to look for clues to tumor DNA in the blood of cancer patients who have not yet developed symptoms, and to screen and diagnose cancer early. To achieve this goal, GRAIL’s clinical trial “Circulating Cell-free Genome Atlas” (CCGA) is one of the largest clinical projects to date. In this trial, GRAIL used a variety of strategies to analyze blood samples from subjects, explore patterns and levels of blood free DNA expression, and help them develop cancer screening products. Last year, the company published the first pre-test data from the CCGA study at the AACR and ESMO annual conferences, showing that its early screening method based on three prototype detection techniques is highly specific in different types of cancer. In addition, at the ASCO annual meeting in June this year, the company’s data showed that its detection method can detect strong signals of 12 deadly cancers in the early stage of cancer, its specificity is at least 99%, and it can identify cancer with high precision. Source of organization. “Most cancers are not discovered until late, and cancer remains the second leading cause of death in the world. To address this challenge, we have launched one of the most ambitious clinical research projects in the field of genomic medicine to support a new pan Early detection methods for cancer,” said Dr. Anne-Renee Hartman, vice president of clinical development at GRAIL. “We are pleased to be able to present the latest data from our large-scale, rigorous clinical research project, which demonstrates that our test method uses a single blood sampling to detect more than 20 types of cancer at various stages and to determine where the cancer is located in the body. Location, while minimizing false positives.”
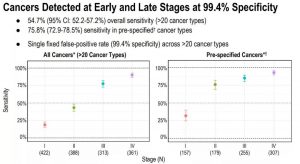
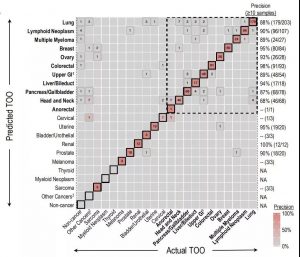
In this multicenter clinical trial, the researchers analyzed cfDNA in 3,583 blood samples, including 1,530 samples of patients diagnosed with cancer and 2,053 samples of cancer-free volunteers. The blood samples of cancer patients cover more than 20 different cancer types, including HR-negative breast cancer, colorectal cancer, esophageal cancer, gallbladder cancer, stomach cancer, head and neck cancer, lung cancer, lymphocytic leukemia, multiple myeloma, ovarian cancer and pancreas. Cancer, etc. The results of the study showed that the overall specificity of the method was 99.4%, which means only 0.6% of false positive results; the overall sensitivity was 54.7% (95% CI: 52.2%-57.2%). For pre-specified high-mortality type cancers (14 cancers that account for approximately 63% of cancer deaths in the United States), the sensitivity of this method is 75.8% (72.9%-78.5%); the sensitivity of stage I cancer patients is 32%, phase II The patient was 76%, the stage III patient was 85%, and the stage IV patient was 93%. In addition, the method can accurately locate the organ or tissue source of the tumor in 97% of the samples, and 89% of the returned results prove to be correct traceability results. This precise traceability of more than 20 different tumor types provides a powerful guarantee for simplifying subsequent diagnosis, demonstrating the broad application prospects of this method in the early screening field.
It is noteworthy that, unlike liquid biopsy methods that detect genetic mutations in DNA or other cancer-related changes, GRAIL’s targeted methylation assay focuses on the analysis of methylation modifications in ctDNA abnormalities. It turns out that in many cases, abnormal patterns of methylation are more indicative of cancer and cancer types than mutations. Dr. Geoffrey Oxnard, the research leader, said: “Our previous work has shown that methylation-based assays are superior to traditional DNA sequencing methods in the detection of multiple forms of cancer using blood samples. This latest study shows that this Detection is a viable way to screen cancer patients.” According to GRAIL, the company’s efficient methylation technology prioritizes the most versatile regions of genomic information and leverages its proprietary database and machine learning algorithms. The presence of cancer is detected and the tissue source of the tumor is identified. In addition, the company’s cancer and non-cancer methylation signature sequencing database is considered to be the largest of its kind, covering approximately 30 million methylation sites throughout the genome, involving more than 20 different stages of cancer. This also provides a strong foundation for the company to develop reliable cancer methylation analysis methods. GRAIL said that in the future, the company will release other new data from its research projects, and the relevant independent verification results will be displayed next month.

